InstaDeep has rolled out its groundbreaking Nucleotide Transformer v3 (NTv3), a multi-species genomics foundation model tailored for 1 MB context lengths at single-nucleotide resolution. Designed to elevate genomic prediction and design, NTv3 bridges the understanding of local DNA motifs with regulatory contexts at mega-base scales. By combining representation learning, functional track prediction, genome annotation, and a powerful generative component, NTv3 promises unprecedented strides in genomics. Featuring U-Net style architecture, pretraining on 9 trillion base pairs, and performance validated across 24 species, this model sets a new benchmark for genomic analytics and sequence generation.
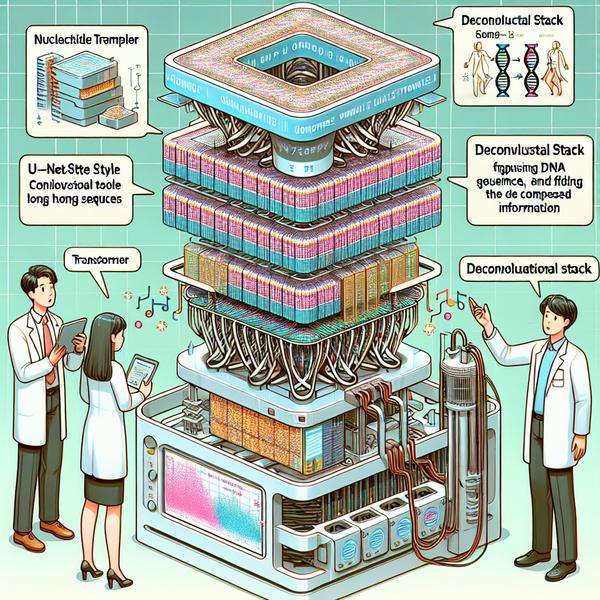
The Power of NTv3's Innovative Architecture
- NTv3's architecture is like building a hierarchical tower that gets detailed as you ascend. It starts with a U-Net style convolutional tower that compresses long DNA sequences into a simpler form. This is similar to shrinking an encyclopedia into a few key phrases but retaining the core ideas.
- Once compressed, the transformer stack steps in. This acts like a detective, finding hidden patterns and long-range relationships within compressed data. Think of it as connecting the dots to see the bigger picture.
- Finally, a deconvolutional tower re-expands this compressed information, providing predictions at single-nucleotide resolution. It’s like zooming back in but with a clearer focus. This triple-step system ensures both efficiency and deep insights.
- Real-world Example: Imagine scanning a beach for shells. First, zoom out for a bird's-eye view, then focus on clusters of rocks, and finally zoom in to examine each shell’s texture. NTv3 does something similar but with genomics.
- The result of this robust multi-layer processing is the generation of highly accurate genomic predictions, which can be further fine-tuned to meet diverse research goals.
The Importance of Multi-Species Training Data
- NTv3 "learnt the language of life" by training on an extensive dataset of 9 trillion base pairs. Just as understanding multiple dialects of a language gives broader perspective, its dataset included over 128,000 species.
- The model also underwent supervised learning, meaning it wasn't just taught to recognize patterns but also how to derive practical insights from them. Picture teaching a robot to recognize cats and then asking it to identify persian cats very specifically —that’s how NTv3 gathers focused precision.
- Cross-species learning means NTv3 now applies its knowledge to both plants and animals. For instance, its understanding from zebrafish data could help predict human gene functions, embodying the phrase, "learning from one tool can sharpen another."
- Real-world Example: Imagine a chef who’s worked with spices from every continent; they’ll be immensely skilled in blending flavors. NTv3 has similarly trained on diverse genomic data to create holistic predictions across species.
- This approach not only widens the model’s usability but ensures adaptability across multiple biological problems, offering unprecedented flexibility for researchers globally.
Breaking Records with NTv3's Benchmark Performance
- NTv3 doesn’t just make promises—it delivers. Its performance was scrutinized through the Ntv3 Benchmark, a comprehensive suite of genomic tasks involving over 106 challenges ranging from cross-species to functional tasks.
- It excelled at both identifying long-range genomic patterns and annotating functions, beating even specialized models. It’s akin to a decathlon champion excelling across all 10 events, proving both depth and breadth of capability.
- One standout feature is its single-nucleotide functionality. This means NTv3 predicts individual DNA "letters" with utmost precision. Picture writing a long story where every letter is perfectly selected — that’s the level of detail here!
- Real-world validation was achieved when NTv3's enhancer predictions were tested in lab conditions (via STARR seq assays), showing accuracy levels more than twice as better as previous methods.
- This consistency establishes NTv3 not just as a long-range genomics powerhouse but a reliable tool for end-to-end biological insights, offering immense value in real-world settings.
From Prediction to Sequence Generation
- NTv3 isn’t merely a tool for observing—it can actively create! Using masked diffusion language modeling, scientists now command NTv3 to fill specific gaps in DNA sequences based on given conditions.
- This is particularly valuable for genomic synthesis. If you wanted a specific DNA sequence to produce more enzyme ‘X,’ NTv3 could design it for you. It's akin to asking an architect to design a room fit for acoustics, and they deliver just what you asked for!
- The Stark Lab experiments showcased over 1,000 new enhancers designed by NTv3. These enhancers performed exactly as intended, proving the model’s ability to "imagine" biology and execute it in labs.
- Real-world Example: Think of enhancing crops to thrive under certain climates or synthesizing molecules for pharmaceutical drugs. NTv3’s capabilities open doors to revolutionary applications in personalized medicine and agricultural science.
- This isn’t just about research; it’s about actively transforming genomics into actionable outputs backed by experimental accuracy, epitomizing the convergence of AI and biotechnology.
Why NTv3 Stands Out from Its Peers
- Compared to other DNA language models like GENA-LM, NTv3 offers a staggering context length of 1 Mb, significantly larger than its counterparts. It’s like reading an entire book instead of just its summary.
- NTv3’s innovative U-Net approach aggregates long genomic contexts, offering a clearer, continuous vision without any information loss. In contrast, other models rely on sparse attention methods which might miss details.
- Unlike other models relying heavily on human genome data, NTv3 has trained with a global perspective, spanning diverse species, giving it a more universal appeal for applications ranging beyond just human health.
- Real-world Example: Consider a language app optimized for one dialect of English versus NTv3, which acts as a global translator covering all dialects. It seamlessly bridges species-specific functionality for better real-world applications.
- This universality combined with supervised and generative capabilities shows why NTv3 is a leap forward, not just an iterative improvement in genomic AI systems.